

Today we’d like to introduce you to Jeffrey C. Silva.
So, before we jump into specific questions about the business, why don’t you give us some details about you and your story.
I am a 1986 graduate of Beverly High School (Beverly, MA), located about 20 miles North East of Boston. My academic interests were math and science as far back as I can remember. English and history were not my strong suites, although my history teacher, Mr. Vogler, had quite an impact on my perspective of higher education and gave me an appreciation for investing in my future education. He played a significant role in changing my trajectory toward college by personally escorting me out of his general history class and into to a college preparatory history class during my sophomore year. I had not done anything wrong, Mr. Vogler simply decided that I did not belong on a general academic track but instead in a college prep program. At the time, I was focused on the inconvenience of the situation, since I would have to work harder to get through the class work. In retrospect, this change provided me with the confidence I needed to push myself through the remainder of high school. More than twenty years later, now retired, Mr. Vogler and I meet for breakfast once or twice a year to catch up on what I am doing and how he is enjoying his retirement, volunteer work and his time golfing.
At the time I graduated high school, I did not know exactly what I wanted to do for a career but knew that if I focused on math and science in my undergraduate schooling, I would be have opportunities presented to me. I attended Merrimack College (in North Andover, MA) for my undergraduate education. I was originally registered as a pre-med student, but eventually decided to major in chemistry with a minor in biochemistry. The professors that I had at Merrimack were outstanding, providing guidance and mentoring that kept their students challenged and motivated their students to strive for excellence. Dr. Pike was a wonderful organic chemistry teacher, a pioneer in micro-scale organic chemistry, and a teacher who cared for his students and was a great role model. Dr. Szafran was an inspiring inorganic chemistry and physical chemistry teacher, who also enlightened his students with applied mathematics in both subjects to sharpen their knowledge and understanding of math principles.
After graduating Merrimack College, I attended the Johns Hopkins University for a Ph.D. program in the Department of Chemistry. I studied natural product biosynthesis in microbial & fungal organisms in the laboratory of Dr. Craig A. Townsend. I was intrigued by the research that was being performed in Dr. Townsend’s laboratory because the work required multidisciplinary scientific training in the areas of chemistry, biochemistry, enzymology and molecular biology. This broad knowledge base was necessary to explore questions related to biosynthesis (creation) of the interesting microbial natural products known as mycotoxins and other secondary metabolites that were developed as antimicrobial therapeutics.
After finishing my Ph.D. at Johns Hopkins, I headed back home to Massachusetts for my post-doctoral training at Harvard Medical School, in the Department of Biological Chemistry and Molecular Pharmacology in the laboratory of Dr. Christopher T. Walsh. I idolized Dr. Walsh during my graduate school years, after taking a class at Johns Hopkins using a book Dr. Walsh authored called Enzymatic Reaction Mechanisms At the end of my graduate study, I became interested in signal transduction, to learn more about how microbes interact with their environment, regulate the myriad of cellular processes required for normal cell growth and differentiation and evolve resistance to antibiotics. The post-doctoral work in Dr. Walsh’s lab allowed me to expand my knowledge in this area.
With my graduate training at Johns Hopkins University in analytical biochemistry and my post-doctoral experience at Harvard Medical School in bacterial signal transduction, I found myself heading toward a career in Proteomics. Proteomics is a multi-disciplinary branch of biotechnology that applies techniques of molecular biology, biochemistry and genetics to analyzing the structure, function and interactions of the proteins in a particular cell, tissue or organism. These proteins are the end product of the cell’s genome. My broad interests in bio-analytical science led me to join a start-up company called Microbia immediately after my post-doc. Microbia’s goal was to bio-engineer microbial organisms to serve as cellular factories for the production of complicated metabolites that could be used as medical therapies or additives in food manufacturing. The bio-engineering was achieved by transferring the yeast genetic information (DNA) of signal transduction machinery into other microbial organisms to reprogram the new microbe to over-produce commercially useful metabolites for the pharmaceutical or food processing industry.
After my work at Microbia (now Ironwood Pharmaceuticals), my interest became focused on establishing new analytical tools that could use to better understand the proteome (protein content) of the new engineered microbials. This ultimately led me to seek an opportunity to focus my scientific training in area of proteomics at a new company called Micromass. At the time I joined Micromass (Manchester, UK) it had a strong reputation as one of the leaders in the development of mass spectrometry (MS) equipment for large-scale protein analysis as well as the software and bioinformatics tools needed to evaluate the data for biological insight. Micromass was acquired by Waters Corporation (Milford, MA) soon after I joined. Waters was known both by academia and by industry as a leader in liquid chromatography (LC) equipment and separation science. While at Waters/Micromass, I was a member a proteomics research and development team that established and commercialized a novel LCMS (liquid chromatography/mass spectrometry) technology called Data Independent Acquisition (DIA), for large-scale protein analysis. I dedicated my time to develop DIA as a brand new comprehensive technology for global protein analysis which can identify and quantify proteins and help better understand the role of these proteins in the cell. The technology was developed as a powerful tool to discover biomarkers for drug discovery and mechanisms of disease biology and could be used to monitor levels of critical proteins in blood for diagnostic applications. My involvement during the research, development and commercialization of this novel mass spectrometry method is represented in numerous patents and publications. The resulting analytical platform was quite versatile and broadly applicable.
My interest in proteomics and signal transduction also led me to focus my efforts on using mass spectrometry to monitor a wide variety of post-translational modifications (PTMs), to help understand the mechanisms of cellular signaling. PTMs are chemical modifications made directly to cellular proteins and are controlled by the cell. PTMs are such an important aspect of Biology because they serve to alter the role, function and location of proteins within the cell and also play a major role cellular differentiation and development. My research efforts led to the development novel strategies that could be used to monitor the status of PTMs on key regulatory proteins within critical cellular signaling networks by using antibodies to capture specific protein components of interest followed by subsequent analysis using standard LCMS methods. These signaling networks are responsible for maintenance of normal cell growth but can also be interrupted to direct cells into a state of uncontrolled growth and lead to multiple forms of cancer and disease progression. I joined Cell Signaling Technology (CST) after leaving Waters Corporation to become the manager of a proteomics service group providing contract research for clients outside of CST, using antibodies to capture proteins from biological samples for subsequent LCMS analysis to compare how these proteins changed between normal and disease (or drug treated) conditions. The contract research group performed hundreds of cutting-edge proteomics projects for the Biotech and Pharmaceutical industry as well as for Academia. At CST, I focused my initial research efforts on application development for monitoring cellular proteins to facilitate drug development, disease biology research and biomarker discovery research. This work lead to many publications demonstrating the use of PTM proteomics for identifying novel biomarkers in a variety of sample types and disease model systems.
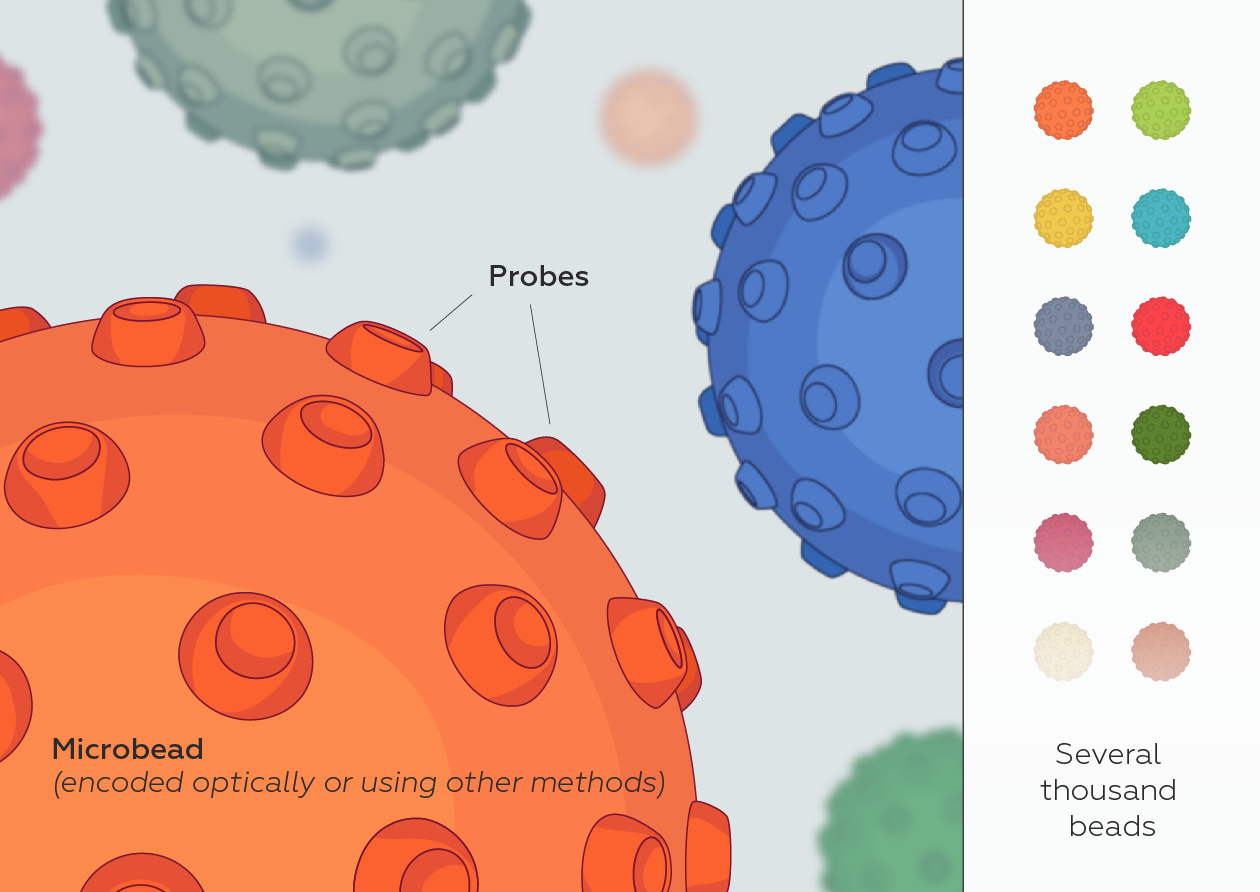
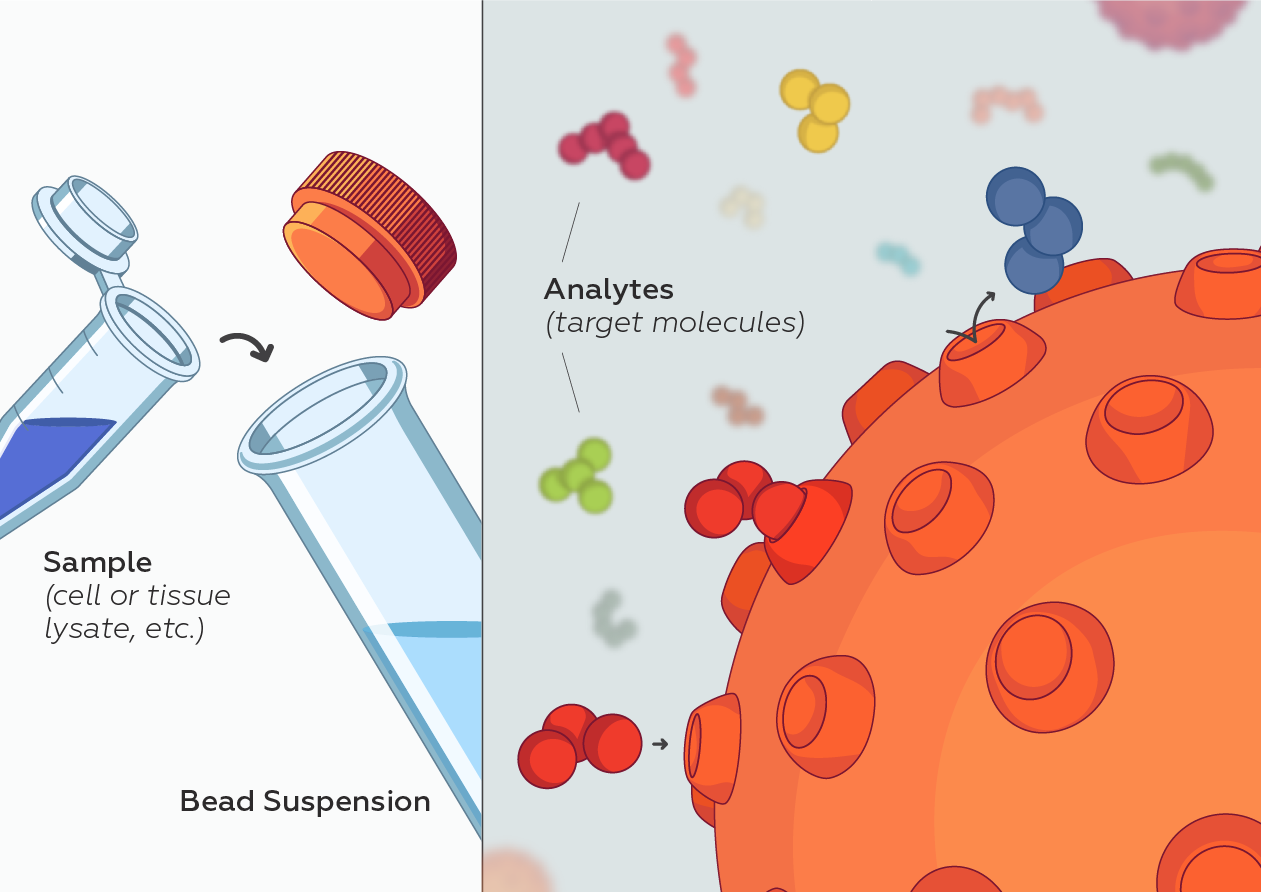
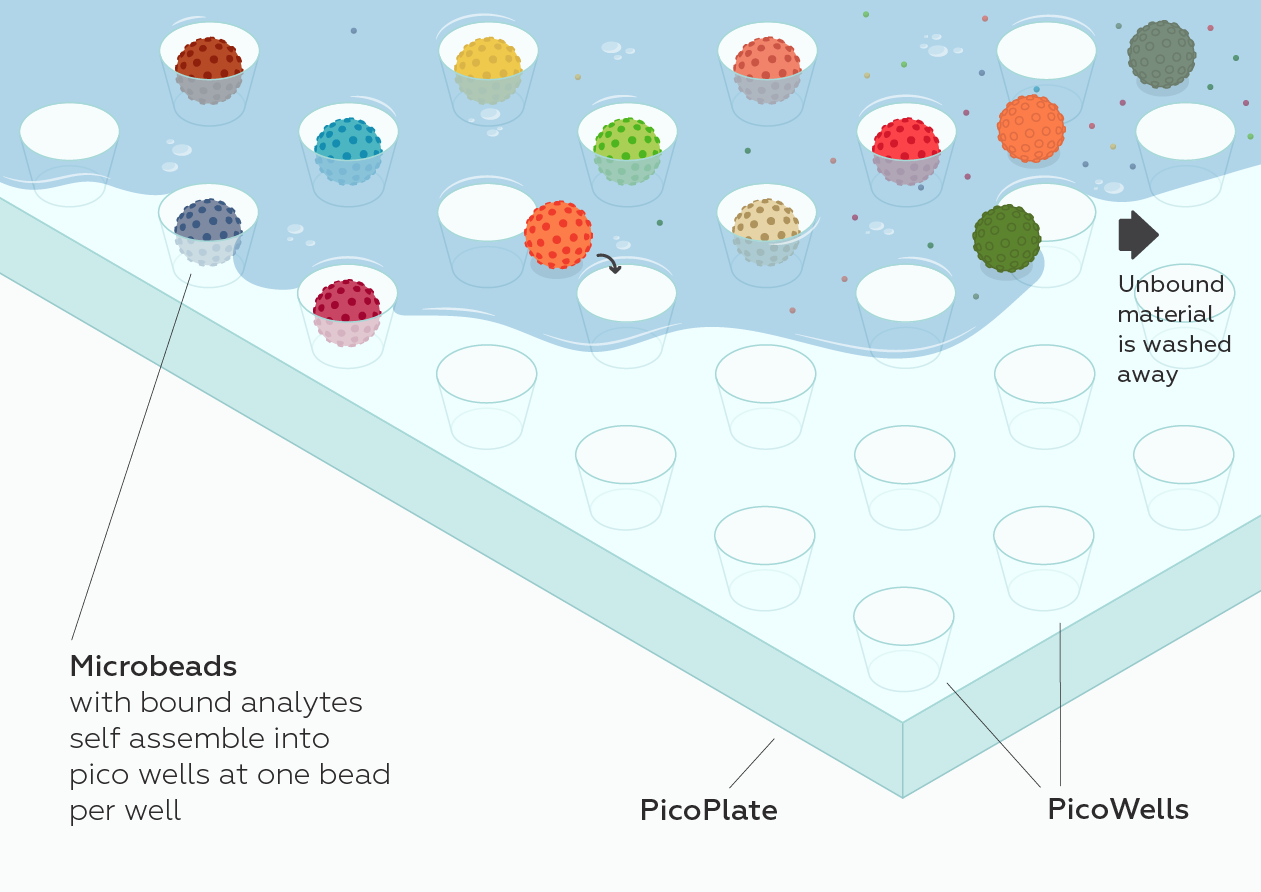
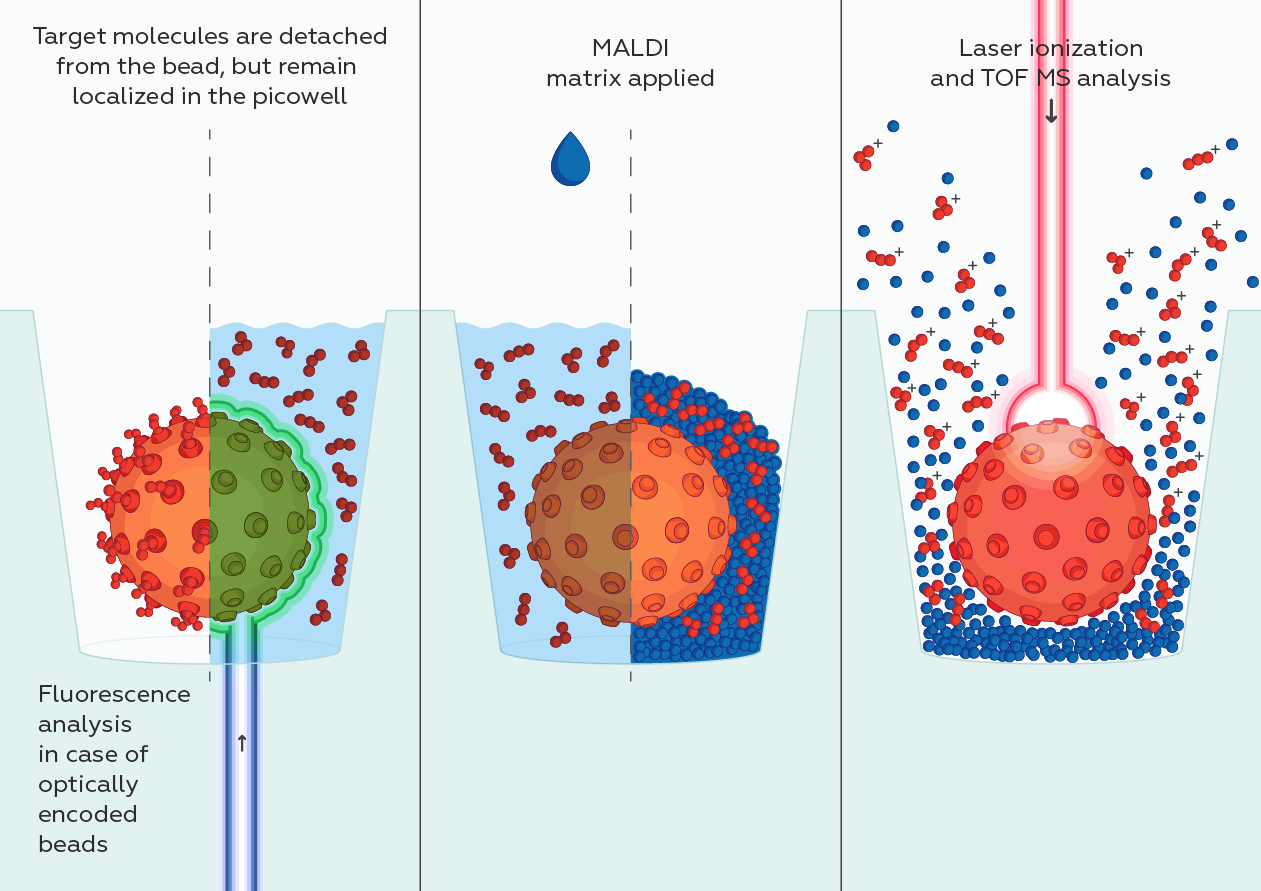
While at CST, starting in 2013, I served as an external industry advisor for North Shore InnoVentures providing advice to Adeptrix for the development of their BAMS (Bead Assisted Mass Spectrometry) technology platform which integrated immunoaffinity enrichment with MALDI mass spectrometry while evaluating the technology platform as a complementary service offering and/or product opportunity for CST. Configuring this type of assay on a MALDI MS instrument would provide a significant improvement over existing LCMS methods by significantly reducing the amount of time it would take to perform each test (allowing many more tests to be performed per day) and is amenable to measuring many more diverse protein and PTMs in a single test (making it an ideal analytical platform for monitoring many different aspects of Biology).
In 2017, I became the Director of Product Development for Adeptrix to help accelerate the development and commercialization of the BAMS technology as a robust bio-analytical platform for targeted proteomics. In the past year, we have built an inventory of approximately 100 protein assays for rapid and efficient measurement of protein biomarkers. Over the 18 months we expect to accumulate an inventory of at least 2000 protein assays that can be used to facilitate research in disease biology and drug development.
A complete list of my published work can be found in My NCBI Bibliography:
http://www.ncbi.nlm.nih.gov/myncbi/browse/collection/48236964/?sort=date&direction=ascending
Great, so let’s dig a little deeper into the story – has it been an easy path overall and if not, what were the challenges you’ve had to overcome?
There have been many challenges throughout my career, that hard work, dedication and perseverance help guide me through to success, however, I would say that the most important challenge in my career is to maintain a balance between good health, family and work (in that order).
Alright – so let’s talk business. Tell us about Adeptrix – what should we know?
Adeptrix is a research tools company that has been established to commercialize a new analytical platform technology termed Bead Assisted Mass Spectrometry (BAMS), to make targeted proteomics widely accessible for Biological research (not just limited to highly specialize proteomic laboratories).
BAMS combines two common technologies: mass spectrometry and immunoassays. Mass spectrometry is currently a low throughput method because it requires extensive upfront sample fractionation by liquid chromatography (LC-MS). Immunoassays such as ELISA and antibody microarrays are low multiplex and are unable to measure hundreds of proteins. BAMS innovatively couples bead-based immunoassays with mass spectrometry to achieve both high multiplex (hundreds of proteins per assay) and high throughput (thousands of assays per day) capability. The BAMS assay platform is an easy-to-use method to perform in-depth proteomic profiling for Life Science research and development.
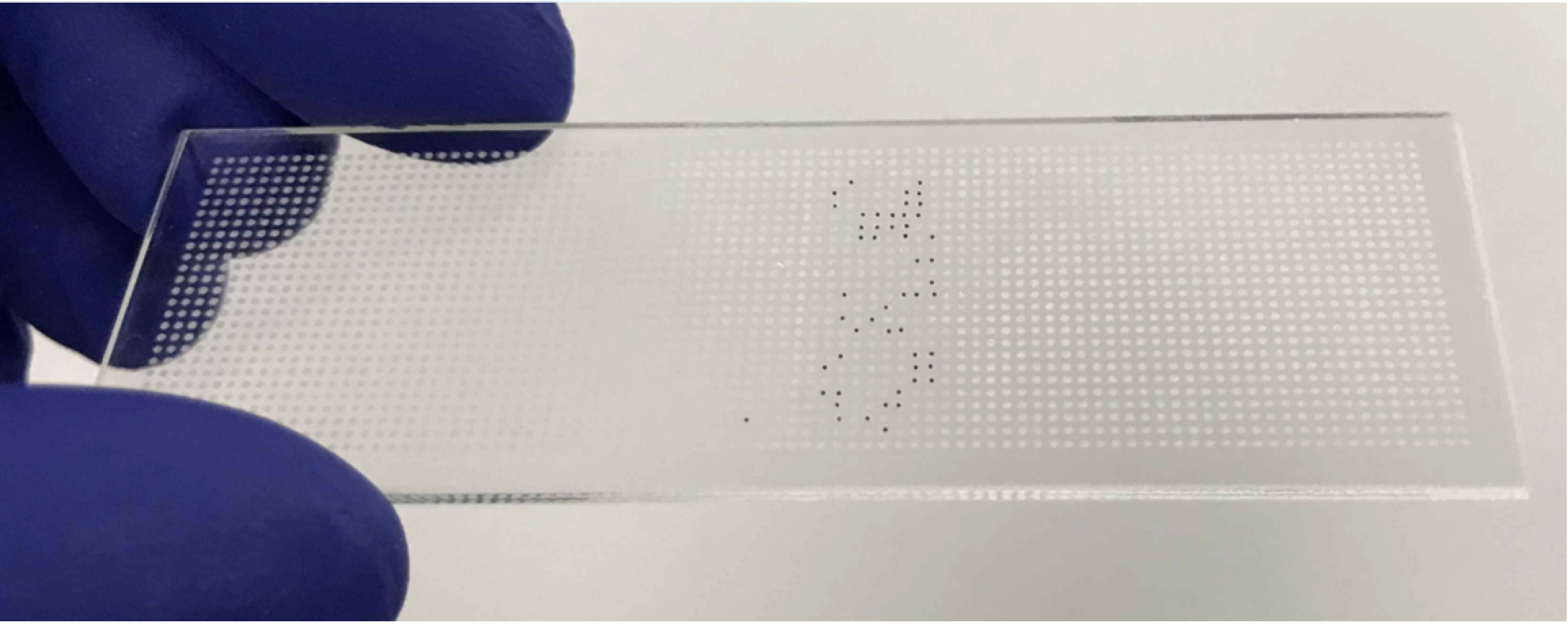
Proteomics is the next frontier (beyond genomics) for discovering new diagnostic and prognostic disease biomarkers. The current proteomic platforms, LC-MS and immunoassays, lack the necessary throughput and/or multiplexing capacity to measure 100’s to 1000’s of protein targets in 100’s to 1000’s of patient samples, which is needed to uncover differences between the diseased and control populations. The BAMS assay platform is a biomarker discovery workflow, which can measure up to 100,000 compounds/day due to its unique miniaturized assay format (the size of a stick of gum), which will dramatically accelerate the current slow pace of protein biomarker discovery research.
Is there a characteristic or quality that you feel is essential to success?
There are many qualities/characteristics that have played an important role in the success and happiness of my career: perseverance to work through the problems that challenge me in my research and product development efforts, curiosity to explore how systems operate, and respect of my colleagues to keep them engaged when approached for advice or insight, genuine excitement for my work.
Contact Info:
- Address: 100 Cummings Center, Suite 438Q
Beverly, MA 01915 - Website: www.adeptrix.com
- Phone: (978) 473-7670
- Email: jsilva.adeptrix@gmail.com
- Other: https://www.linkedin.com/in/jeffreycruzsilva/


Getting in touch: BostonVoyager is built on recommendations from the community; it’s how we uncover hidden gems, so if you know someone who deserves recognition please let us know here.
















